Current Questions
How do cells organize their genomes?
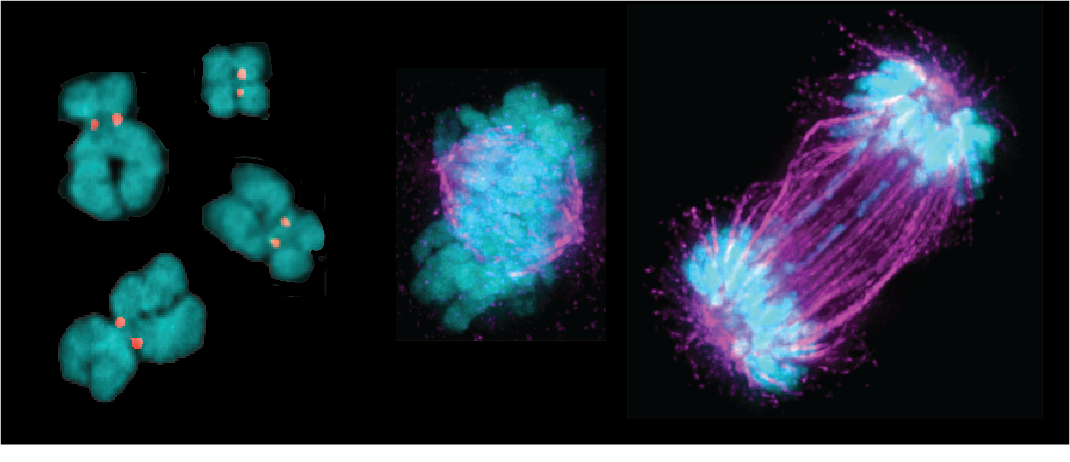
The cohesin complex is a key regulator of genome integrity, contributing to multiple functions including DNA organization, DNA replication, transcription, the DNA damage response, and cell division. But how does the cohesin complex know where to go and what job to do?
We are using large-scale screening approaches combined with targeted genetic and molecular techniques to identify regulators of the cohesin complex and define their function. Using this approach, we recently discovered the cohesin regulatory protein PRR12. Current work seeks to understand the role of PRR12 in regulating cohesin stability and the DNA damage response.
Publications
Nguyen, AL., Smith, EM., Cheeseman, IM. Developmental Cell (2025)
How is cell division altered across cellular contexts?
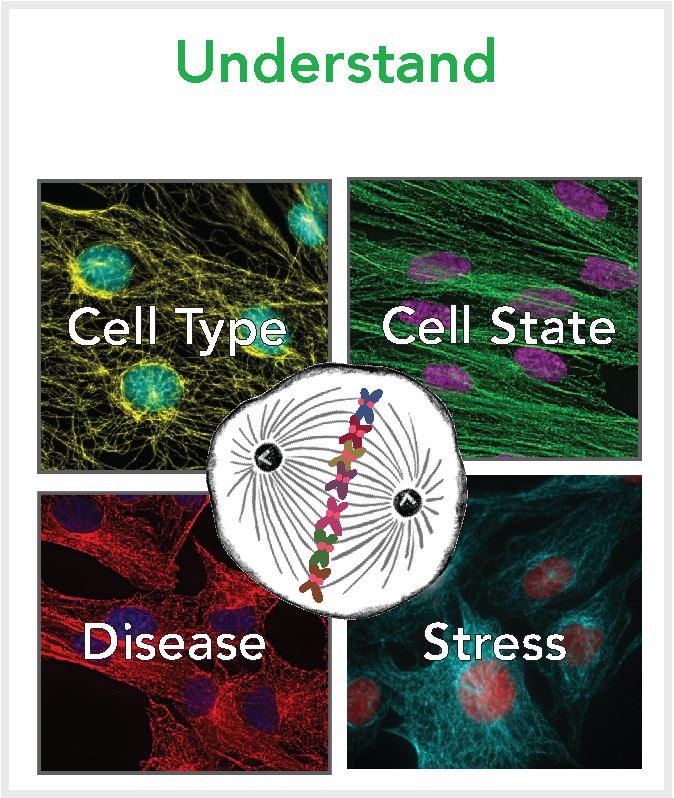
Cell division is a fundamental process where cells duplicate and segregate their genomes. To achieve this, hundreds of proteins must function together in a carefully choreographed dance to ensure the chromosomes are segregated faithfully. We are fascinated in understanding how these mechanisms are rewired across diverse cellular contexts including in different cell types, cells states, in stress conditions, and within disease.
By harnessing differential gene requirements we uncovered the function of the enigmatic CENP-O complex in cell division and identified why these proteins are only required in select cancers. Current work builds on these approaches, investigating other selectively required cell division genes. We seek to define the function of these proteins, identify why they are selectively required, and determine how we can exploit these factors to selectively target disease.
Publications
Nguyen, AL., Fadel, MD., Cheeseman, IM. Molecular Biology of the Cell (2021)
How do gene requirements change across species?
Mice and other mammals are often used as models for human disease, with the assumption that the proteins and pathways in these organisms will be highly conserved. However our work and others has begun to reveal striking differences in gene requirements across even closely related mammalian species. Understanding these differences will help reveal the fundamental biology by which cells function and provide critical insight into the evolution of core cellular processes.
To achieve this, we are performing pooled CRIPSR/CAS9 functional genetic screens in cell lines from diverse animal models.